Bayesian/Frequentist Tutorial¶
[1]:
import arviz as az
import bambi as bmb
import numpy as np
import pandas as pd
import pymc3 as pm
import matplotlib.pyplot as plt
from pymer4.simulate import simulate_lm, simulate_lmm
from pymer4.models import Lmer, Lm
from scipy.stats import ttest_ind
/home/osvaldo/anaconda3/lib/python3.7/site-packages/rpy2/robjects/pandas2ri.py:14: FutureWarning: pandas.core.index is deprecated and will be removed in a future version. The public classes are available in the top-level namespace.
from pandas.core.index import Index as PandasIndex
INFO:numexpr.utils:NumExpr defaulting to 4 threads.
[2]:
az.style.use('arviz-darkgrid')
Generate t-test data¶
[3]:
a = np.random.normal(5,2,1000)
b = np.random.normal(8,2.5,1000)
df = pd.DataFrame({'Group':['a']*1000 + ['b']*1000,'Val':np.hstack([a,b])})
[4]:
df.groupby('Group').describe()
[4]:
| Val | ||||||||
|---|---|---|---|---|---|---|---|---|
| count | mean | std | min | 25% | 50% | 75% | max | |
| Group | ||||||||
| a | 1000.0 | 4.901747 | 1.955774 | -0.900373 | 3.586848 | 4.961405 | 6.163554 | 11.872290 |
| b | 1000.0 | 7.975125 | 2.518749 | -2.232115 | 6.278875 | 8.002316 | 9.669793 | 14.847826 |
[5]:
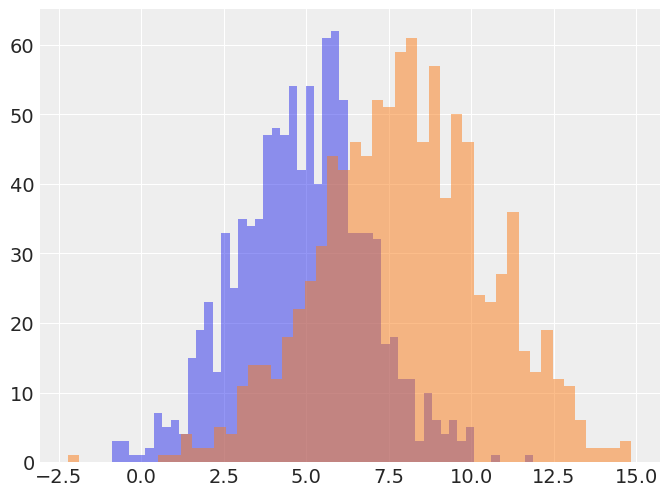
f,ax = plt.subplots(1,1,figsize=(8,6))
ax.hist(a,alpha=.5,bins=50);
ax.hist(b,alpha=.5,bins=50);
Frequentist¶
Since this analysis is relateively straightforward we can perform a between groups t-test using scipy
[6]:
ttest_ind(b,a)
[6]:
Ttest_indResult(statistic=30.477105041971846, pvalue=7.2541984106044045e-168)
We can also set this up as a dummy-coded univariate regression model which is identical
[7]:
# Using the pymer4 package, but we could have used statsmodels instead
model = Lm('Val ~ Group',data=df)
model.fit()
Formula: Val~Group
Family: gaussian Estimator: OLS
Std-errors: non-robust CIs: standard 95% Inference: parametric
Number of observations: 2000 R^2: 0.317 R^2_adj: 0.317
Log-likelihood: -4463.088 AIC: 8930.176 BIC: 8941.378
Fixed effects:
[7]:
| Estimate | 2.5_ci | 97.5_ci | SE | DF | T-stat | P-val | Sig | |
|---|---|---|---|---|---|---|---|---|
| Intercept | 4.902 | 4.762 | 5.042 | 0.071 | 1998 | 68.742 | 0.0 | *** |
| Group[T.b] | 3.073 | 2.876 | 3.271 | 0.101 | 1998 | 30.477 | 0.0 | *** |
Bayesian¶
We can compute the equivalent dummy-coded regression model to estimate with bambi and the pymc3 backend
[8]:
b_model = bmb.Model(df)
res_b = b_model.fit('Val ~ Group', samples=1000, tune=1000)
Auto-assigning NUTS sampler...
INFO:pymc3:Auto-assigning NUTS sampler...
Initializing NUTS using jitter+adapt_diag...
INFO:pymc3:Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (2 chains in 2 jobs)
INFO:pymc3:Multiprocess sampling (2 chains in 2 jobs)
NUTS: [Val_sd, Group, Intercept]
INFO:pymc3:NUTS: [Val_sd, Group, Intercept]
[9]:
# Model priors
b_model.plot();
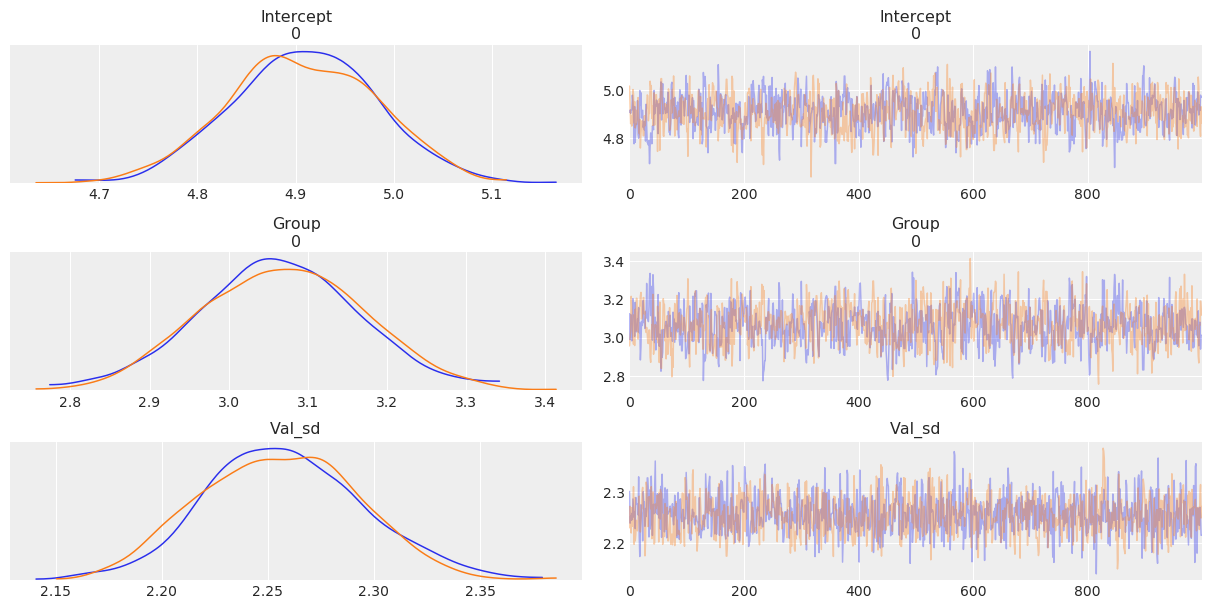
[10]:
az.plot_trace(res_b);
[11]:
az.summary(res_b)
[11]:
| mean | sd | hpd_3% | hpd_97% | mcse_mean | mcse_sd | ess_mean | ess_sd | ess_bulk | ess_tail | r_hat | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Intercept[0] | 4.908 | 0.073 | 4.775 | 5.047 | 0.003 | 0.002 | 780.0 | 780.0 | 788.0 | 1197.0 | 1.0 |
| Group[0] | 3.066 | 0.101 | 2.866 | 3.245 | 0.004 | 0.003 | 723.0 | 717.0 | 726.0 | 1067.0 | 1.0 |
| Val_sd | 2.257 | 0.037 | 2.193 | 2.329 | 0.001 | 0.001 | 1549.0 | 1545.0 | 1563.0 | 1408.0 | 1.0 |
[12]:
# Grab just the posterior of the term of interest (group)
group_posterior = res_b.posterior['Group'].values
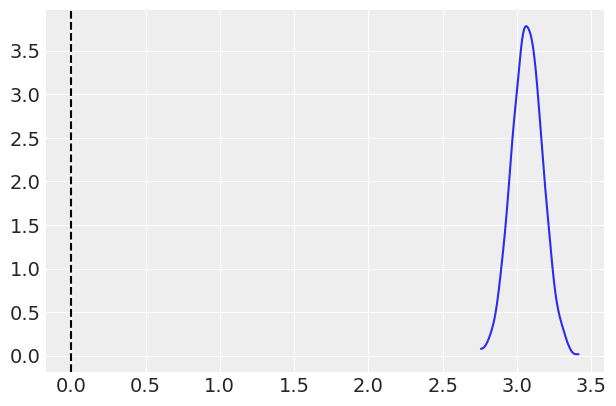
ax = az.plot_kde(group_posterior)
ax.axvline(0, 0, 3, linestyle='--', color='k');
[13]:
#Probabiliy that posterior is > 0
(group_posterior > 0).mean()
[13]:
1.0
Generate multi-level regression data¶
Generate data for a multivariate regression model with random intercepts and slope effect for each group
[14]:
# Simulate some multi-level data with pymer4
df, blups, coefs = simulate_lmm(num_obs=500, num_coef=2, num_grps=25, coef_vals=[5,3,-1])
df.head()
blups.head()
[14]:
| Intercept | IV1 | IV2 | |
|---|---|---|---|
| Grp1 | 5.078462 | 2.965038 | -0.809101 |
| Grp2 | 5.038959 | 3.389151 | -0.728827 |
| Grp3 | 4.711509 | 2.892428 | -1.009656 |
| Grp4 | 4.916194 | 2.633570 | -0.881529 |
| Grp5 | 5.069100 | 3.195954 | -1.023484 |
Frequentist multi-level model¶
[15]:
# Fit multi-level model using pymer4 (lmer in R)
model = Lmer('DV ~ IV1 + IV2 + (IV1 + IV2|Group)',data=df)
model.fit()
Model failed to converge with max|grad| = 0.0024797 (tol = 0.002, component 1)
Formula: DV~IV1+IV2+(IV1+IV2|Group)
Family: gaussian Inference: parametric
Number of observations: 12500 Groups: {'Group': 25.0}
Log-likelihood: -17857.782 AIC: 35715.565
Random effects:
Name Var Std
Group (Intercept) 0.057 0.238
Group IV1 0.081 0.285
Group IV2 0.055 0.235
Residual 0.998 0.999
IV1 IV2 Corr
Group (Intercept) IV1 -0.020
Group (Intercept) IV2 0.149
Group IV1 IV2 -0.115
Fixed effects:
[15]:
| Estimate | 2.5_ci | 97.5_ci | SE | DF | T-stat | P-val | Sig | |
|---|---|---|---|---|---|---|---|---|
| (Intercept) | 5.008 | 4.913 | 5.103 | 0.048 | 24.002 | 103.450 | 0.0 | *** |
| IV1 | 3.015 | 2.902 | 3.128 | 0.058 | 24.031 | 52.268 | 0.0 | *** |
| IV2 | -0.897 | -0.991 | -0.803 | 0.048 | 24.000 | -18.706 | 0.0 | *** |
[16]:
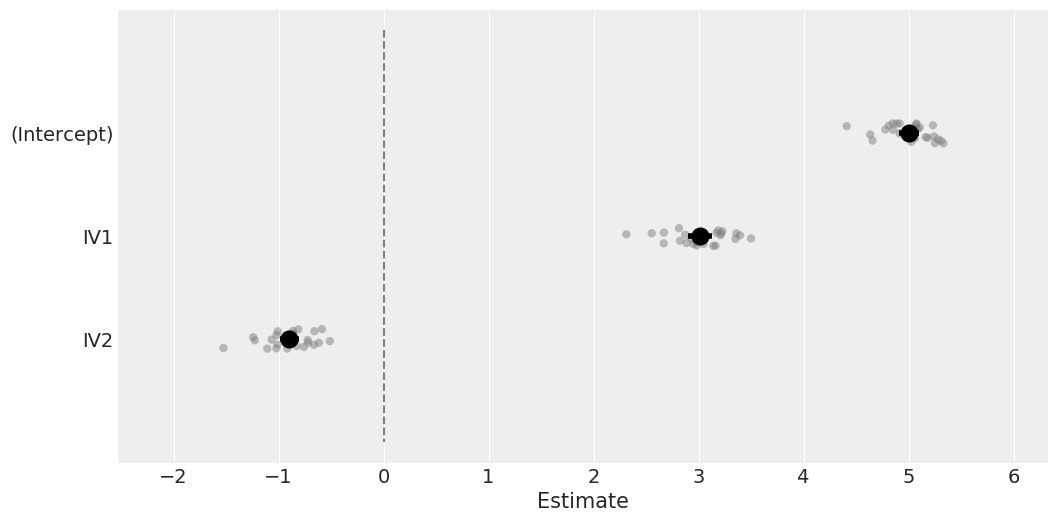
# Plot coefficients and the group BLUPs as well
model.plot_summary();
[17]:
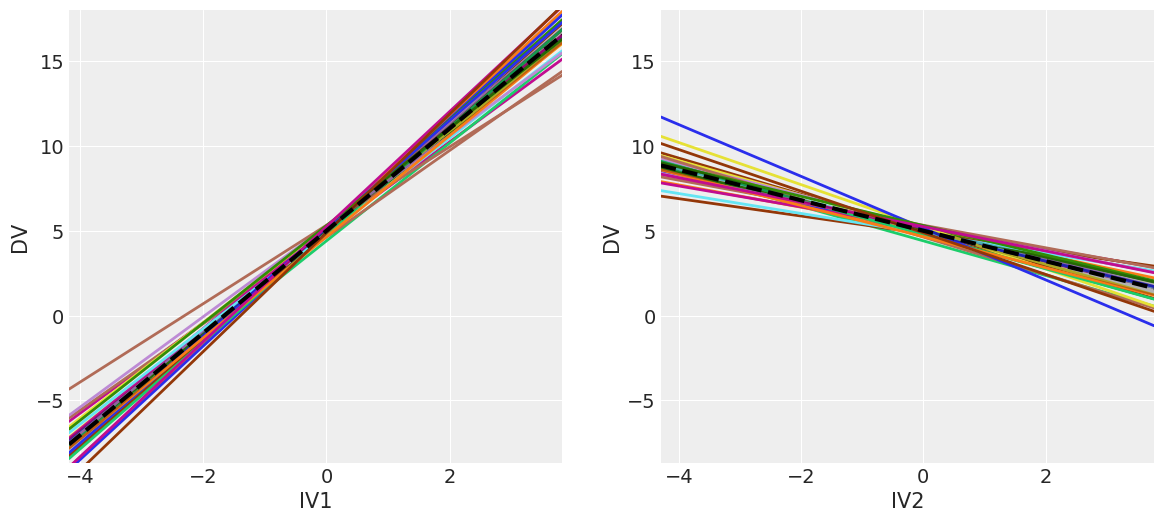
# Alternatively visualize coefficients as regression lines with BLUPs overlaid
_, axs = plt.subplots(1, 2, figsize=(14, 6))
model.plot('IV1', ax=axs[0],)
model.plot('IV2', ax=axs[1]);
Bayesian multi-level model¶
[18]:
b_model = bmb.Model(df)
results = b_model.fit('DV ~ IV1 + IV2',random=['IV1+IV2|Group'], samples=1000, tune=1000)
Auto-assigning NUTS sampler...
INFO:pymc3:Auto-assigning NUTS sampler...
Initializing NUTS using jitter+adapt_diag...
INFO:pymc3:Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (2 chains in 2 jobs)
INFO:pymc3:Multiprocess sampling (2 chains in 2 jobs)
NUTS: [DV_sd, IV2|Group_offset, IV2|Group_sd, IV1|Group_offset, IV1|Group_sd, 1|Group_offset, 1|Group_sd, IV2, IV1, Intercept]
INFO:pymc3:NUTS: [DV_sd, IV2|Group_offset, IV2|Group_sd, IV1|Group_offset, IV1|Group_sd, 1|Group_offset, 1|Group_sd, IV2, IV1, Intercept]
The estimated number of effective samples is smaller than 200 for some parameters.
ERROR:pymc3:The estimated number of effective samples is smaller than 200 for some parameters.
[19]:
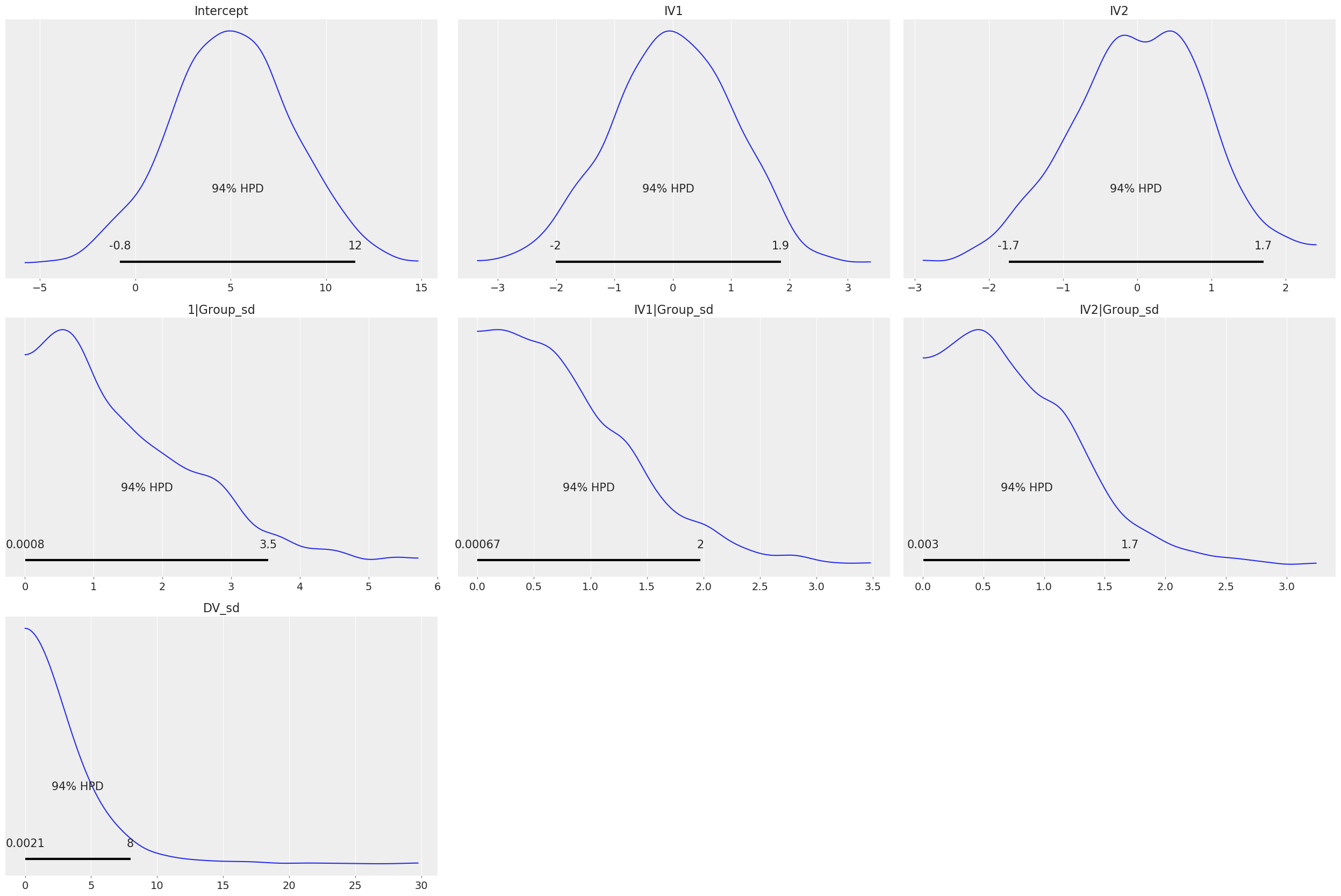
# Plot priors
b_model.plot();
[20]:
#Plot posteriors
az.plot_trace(results, compact=True);
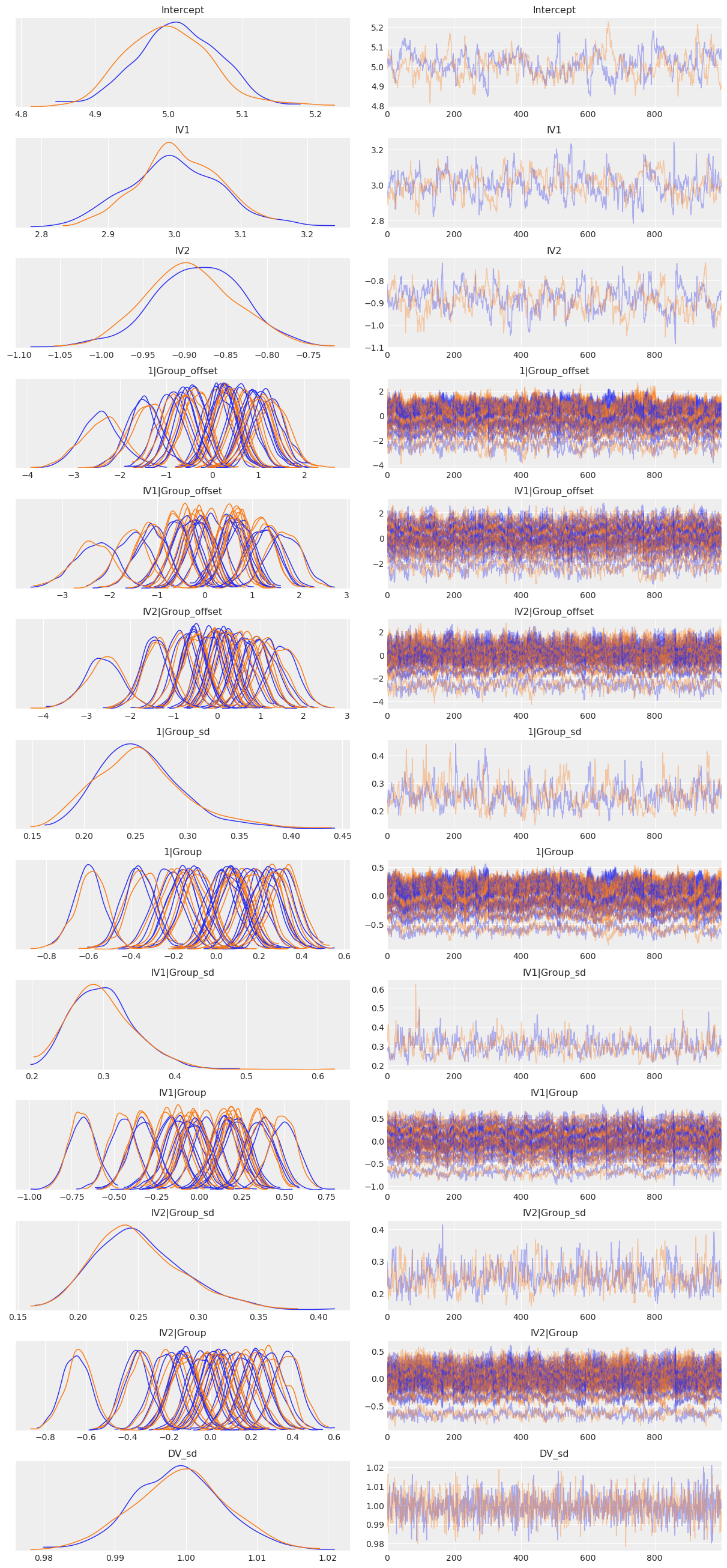
[21]:
az.summary(results,
var_names=['Intercept', 'IV1', 'IV2', '1|Group_sd', 'IV1|Group_sd', 'IV2|Group_sd', 'DV_sd'])
[21]:
| mean | sd | hpd_3% | hpd_97% | mcse_mean | mcse_sd | ess_mean | ess_sd | ess_bulk | ess_tail | r_hat | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Intercept[0] | 5.007 | 0.059 | 4.903 | 5.117 | 0.005 | 0.003 | 142.0 | 141.0 | 143.0 | 256.0 | 1.01 |
| IV1[0] | 3.003 | 0.067 | 2.885 | 3.137 | 0.005 | 0.004 | 160.0 | 160.0 | 159.0 | 397.0 | 1.01 |
| IV2[0] | -0.888 | 0.055 | -0.988 | -0.779 | 0.005 | 0.003 | 146.0 | 144.0 | 146.0 | 298.0 | 1.01 |
| 1|Group_sd | 0.255 | 0.043 | 0.175 | 0.334 | 0.003 | 0.002 | 221.0 | 221.0 | 216.0 | 457.0 | 1.00 |
| IV1|Group_sd | 0.300 | 0.045 | 0.218 | 0.382 | 0.003 | 0.002 | 314.0 | 314.0 | 309.0 | 622.0 | 1.01 |
| IV2|Group_sd | 0.251 | 0.038 | 0.183 | 0.322 | 0.002 | 0.001 | 322.0 | 322.0 | 319.0 | 559.0 | 1.00 |
| DV_sd | 0.999 | 0.007 | 0.988 | 1.012 | 0.000 | 0.000 | 1919.0 | 1919.0 | 1917.0 | 1526.0 | 1.00 |
[22]:
az.plot_kde(results.posterior['IV1'], plot_kwargs={'color': 'C0'}, label='IV1')
az.plot_kde(results.posterior['IV2'], plot_kwargs={'color': 'C1'}, label='IV2')
az.plot_kde(results.posterior['Intercept'], plot_kwargs={'color': 'C2'}, label='Intercept')
plt.axvline(0, color='k', linestyle='--')
plt.legend();
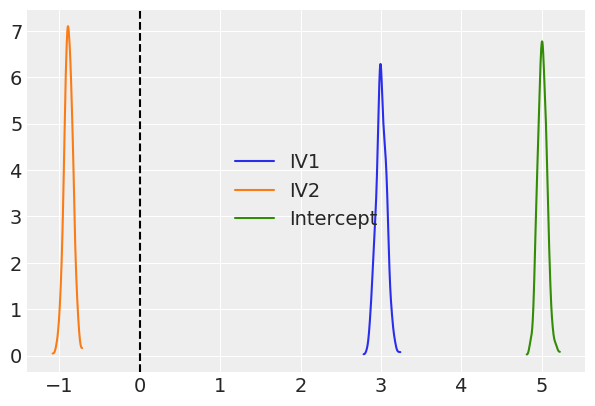
Because we used pymc3 for the backend estimation in bambi, we have access to few extra goodies. Here we make a forest plot similar to the one above for the frequentist model, but with 94% credible intervals instead
[23]:
# Credible interval plot using pymc3
# Line is 94% credible interval calculated as higher posterior density
# Inter quartile range is thicker line
# Dot is median
az.plot_forest(results, var_names=['Intercept', 'IV1', 'IV2', '1|Group_sd'], figsize=(10, 2));
INFO:numba.transforms:finding looplift candidates
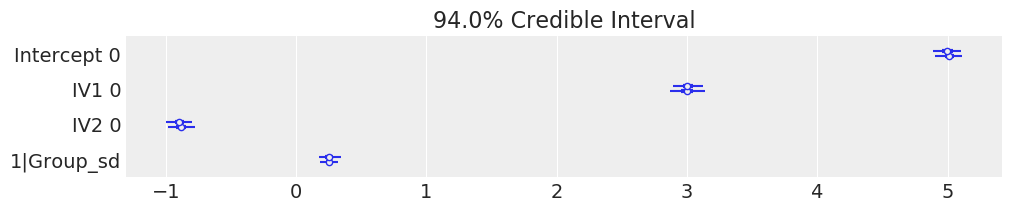
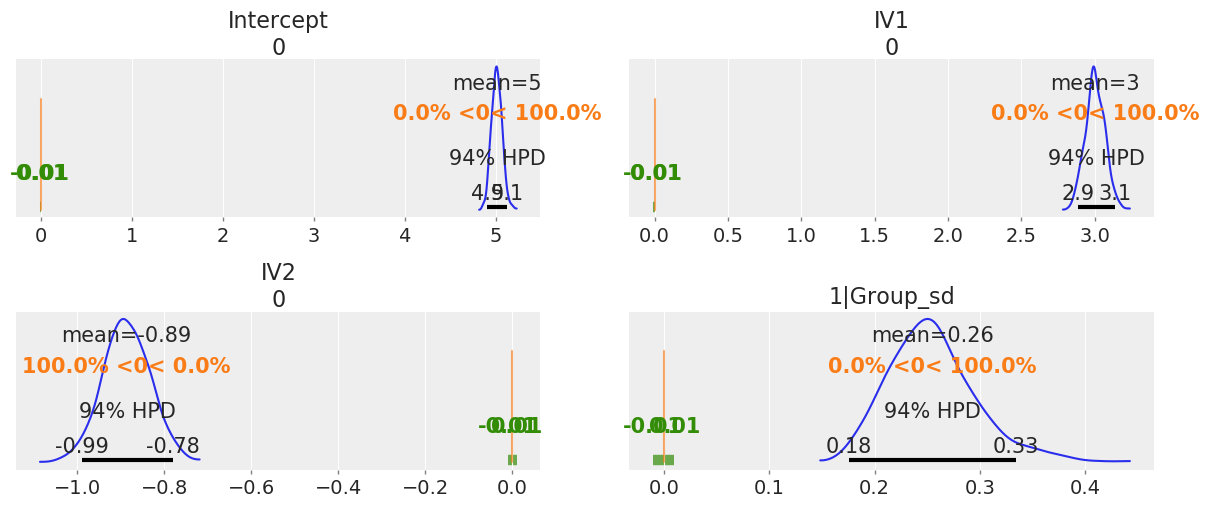
We can also plot the posterior overlayed with a region of practical equivalence (ROPE), i.e. range of values that were the coefficients to fall into, we might interpret them differently. We can see that all our posterior distributions fall outside of this range.
[24]:
# Show credible interval cutoffs, and also overlay region of practical equivalence (arbitrary, in this case close enough to 0 to not matter)
_, ax = plt.subplots(2, 2, figsize=(12, 5), constrained_layout=True)
az.plot_posterior(b_model.backend.trace,
var_names=['Intercept', 'IV1', 'IV2', '1|Group_sd'],
ref_val=0,
rope=[-.01, .01],
ax=ax);